Latent Diffusion Autoencoders: Toward Efficient and Meaningful Unsupervised Representation Learning in Medical Imaging
Abstract
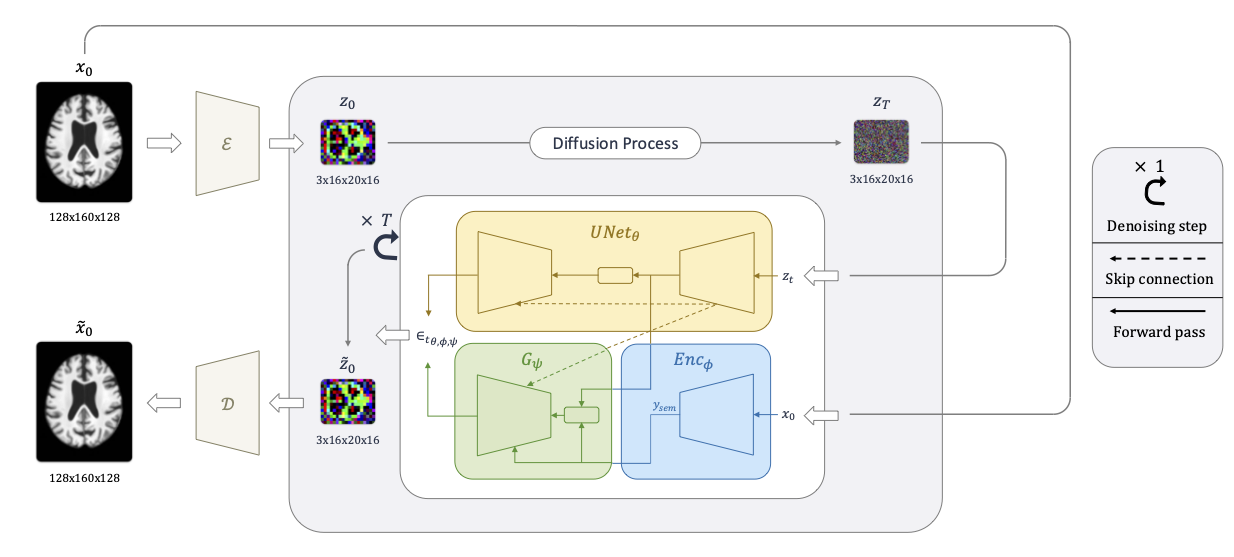
This study presents Latent Diffusion Autoencoder (LDAE), a novel encoder-decoder diffusion-based framework for efficient and meaningful unsupervised learning in medical imaging, focusing on Alzheimer disease (AD) using brain MR from the ADNI database as a case study. Unlike conventional diffusion autoencoders operating in image space, LDAE applies the diffusion process in a compressed latent representation, improving computational efficiency and making 3D medical imaging representation learning tractable. To validate the proposed approach, we explore two key hypotheses: (i) LDAE effectively captures meaningful semantic representations on 3D brain MR associated with AD and ageing, and (ii) LDAE achieves high-quality image generation and reconstruction while being computationally efficient. Experimental results support both hypotheses: (i) linear-probe evaluations demonstrate promising diagnostic performance for AD (ROC-AUC: 90%, ACC: 84%) and age prediction (MAE: 4.1 years, RMSE: 5.2 years); (ii) the learned semantic representations enable attribute manipulation, yielding anatomically plausible modifications; (iii) semantic interpolation experiments show strong reconstruction of missing scans, with SSIM of 0.969 (MSE: 0.0019) for a 6-month gap. Even for longer gaps (24 months), the model maintains robust performance (SSIM > 0.93, MSE < 0.004), indicating an ability to capture temporal progression trends; (iv) compared to conventional diffusion autoencoders, LDAE significantly increases inference throughput (20x faster) while also enhancing reconstruction quality. These findings position LDAE as a promising framework for scalable medical imaging applications, with the potential to serve as a foundation model for medical image analysis. Code available at https://github.com/GabrieleLozupone/LDAE
Community
I'm excited to share our work on Latent Diffusion Autoencoders (LDAE) for unsupervised representation learning in 3D medical imaging. This framework addresses critical challenges in medical AI:
It achieves 20x faster inference than voxel-space approaches while improving reconstruction fidelity
It enables semantic control of clinically meaningful features (e.g., manipulating Alzheimer's atrophy patterns)
It produces high-quality representations for downstream tasks like classification (89.48% ROC-AUC) and age prediction (4.16 years MAE)
Our approach combines AutoencoderKL for compression with a diffusion model in latent space guided by a semantic encoder. This enables not just reconstruction but meaningful manipulation of brain MRI features without requiring labeled data.
I'd appreciate discussions on:
Applications in other medical imaging domains
Potential clinical use cases for semantic manipulation
Ideas for further improving disentanglement in the semantic space
Code is available at our GitHub repo linked in the paper. Looking forward to your insights!
This is an automated message from the Librarian Bot. I found the following papers similar to this paper.
The following papers were recommended by the Semantic Scholar API
- Identity Preserving Latent Diffusion for Brain Aging Modeling (2025)
- TIDE : Temporal-Aware Sparse Autoencoders for Interpretable Diffusion Transformers in Image Generation (2025)
- MoEDiff-SR: Mixture of Experts-Guided Diffusion Model for Region-Adaptive MRI Super-Resolution (2025)
- MAD-AD: Masked Diffusion for Unsupervised Brain Anomaly Detection (2025)
- USP: Unified Self-Supervised Pretraining for Image Generation and Understanding (2025)
- Resolution Invariant Autoencoder (2025)
- REGEN: Learning Compact Video Embedding with (Re-)Generative Decoder (2025)
Please give a thumbs up to this comment if you found it helpful!
If you want recommendations for any Paper on Hugging Face checkout this Space
You can directly ask Librarian Bot for paper recommendations by tagging it in a comment:
@librarian-bot
recommend
Models citing this paper 0
No model linking this paper
Datasets citing this paper 0
No dataset linking this paper
Spaces citing this paper 0
No Space linking this paper