Update README.md
Browse files
README.md
CHANGED
|
@@ -9,8 +9,7 @@ datasets:
|
|
| 9 |
|
| 10 |
## Model description
|
| 11 |
|
| 12 |
-
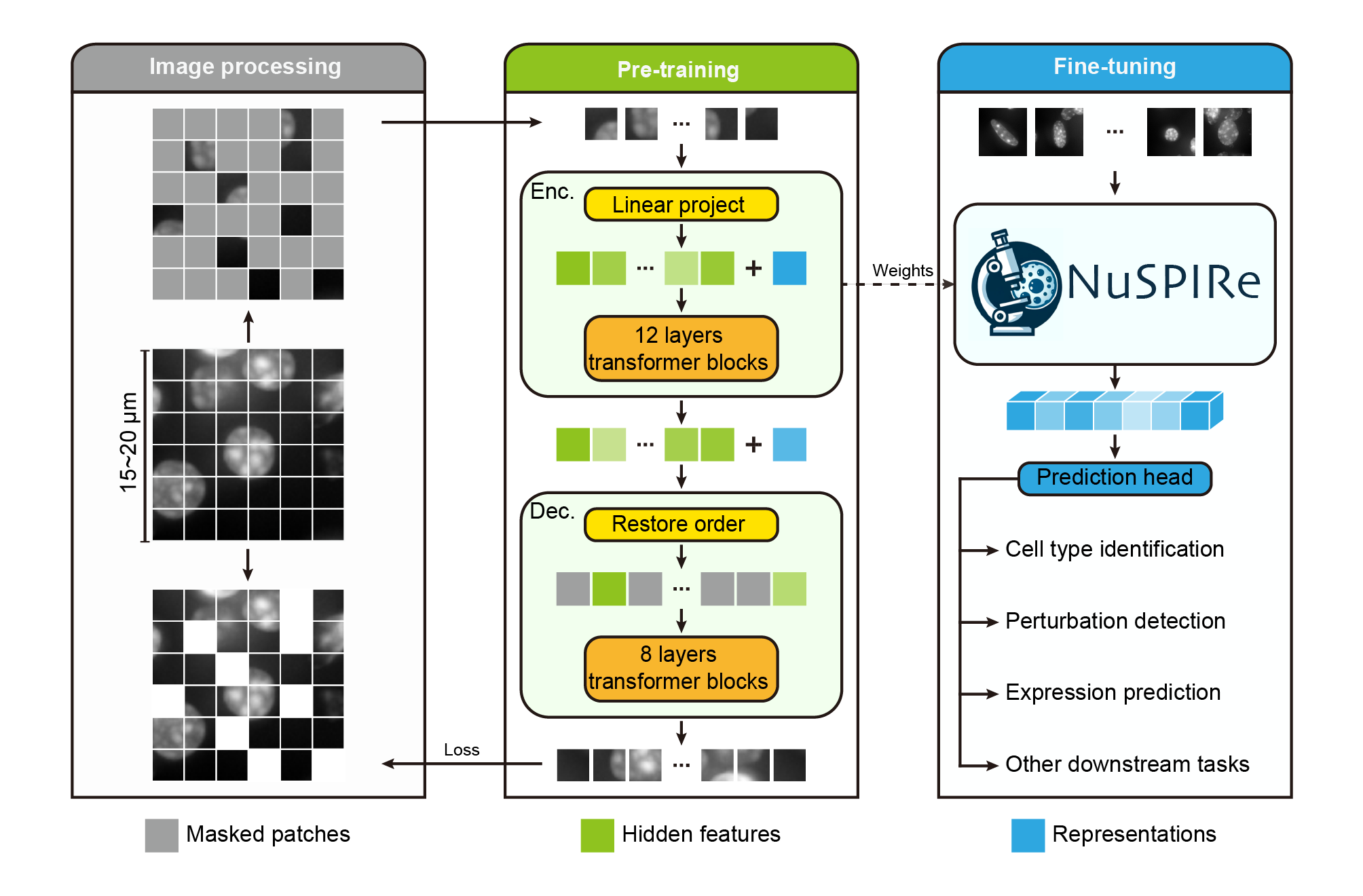
NuSPIRe (Nuclear Morphology focused Self-supervised Pretrained model for Image Representations) is a deep learning
|
| 13 |
-
|
| 14 |
|
| 15 |
|
| 16 |
## Training Details
|
|
@@ -133,9 +132,10 @@ print("Logits:", logits)
|
|
| 133 |
print("Loss:", loss.item())
|
| 134 |
|
| 135 |
```
|
| 136 |
-
|
| 137 |
## Citation
|
| 138 |
|
| 139 |
If you use NuSPIRe in your research, please cite the following paper:
|
| 140 |
|
| 141 |
-
Hua, Y., Li, S., & Zhang, Y. (2024). NuSPIRe: Nuclear Morphology focused Self-supervised Pretrained model for Image Representations.
|
|
|
|
|
|
| 9 |
|
| 10 |
## Model description
|
| 11 |
|
| 12 |
+
NuSPIRe (Nuclear Morphology focused Self-supervised Pretrained model for Image Representations) is a deep learning framework designed to extract nuclear morphological features from DAPI-stained images for guiding field-of-view (FOV) optimization in spatial omics. Trained using self-supervised learning on 15.52 million unlabeled nuclear images from diverse tissues, NuSPIRe leverages pre-existing imaging data to identify biologically informative regions. While primarily developed for FOV selection and layout refinement, it also offers potential for broader morphology-based spatial inference.
|
|
|
|
| 13 |

|
| 14 |
|
| 15 |
## Training Details
|
|
|
|
| 132 |
print("Loss:", loss.item())
|
| 133 |
|
| 134 |
```
|
| 135 |
+
<!--
|
| 136 |
## Citation
|
| 137 |
|
| 138 |
If you use NuSPIRe in your research, please cite the following paper:
|
| 139 |
|
| 140 |
+
Hua, Y., Li, S., & Zhang, Y. (2024). NuSPIRe: Nuclear Morphology focused Self-supervised Pretrained model for Image Representations.
|
| 141 |
+
-->
|