The dataset viewer is not available because its heuristics could not detect any supported data files. You can try uploading some data files, or configuring the data files location manually.
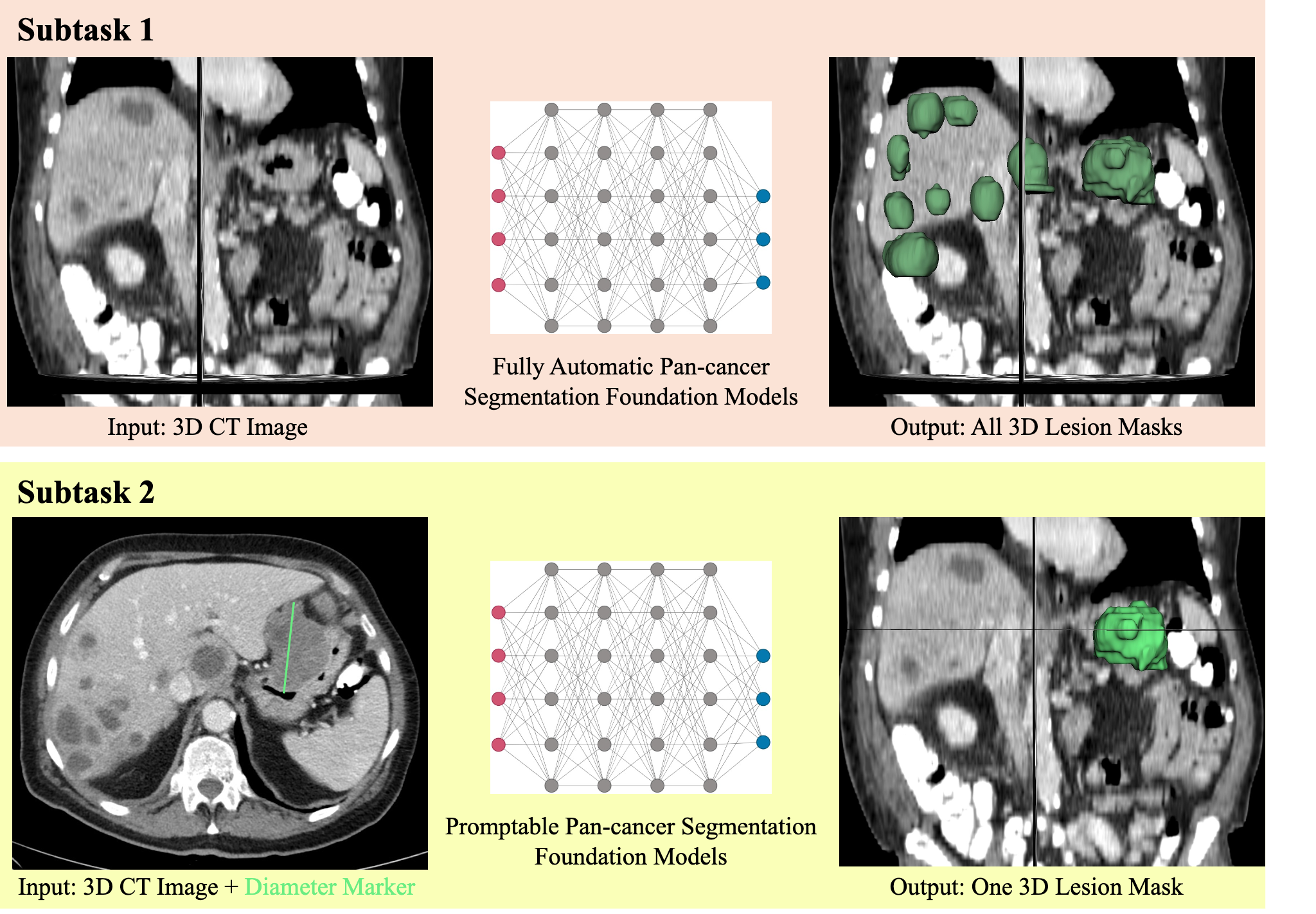
MICCAI FLARE 2025 Pan-cancer Segmentation Subtask 2 (Homepage)
RECIST has been the widely used measurement to quantify the lesion, which use 2D diameter on the largest lesion slice to approximate the lesion size. However, the diameter cannot capture the complete lesion 3D morphology. This subtask aims to develop lightweight segmentation models to produce 3D lesion masks based on 2D RECIST annotation (diameter marker) on laptop (without using GPU during inference).
- input: 3D image (npz format) and RECIST annotation (diameter marker)
- output: 3D lesion segmentation mask
To avoid the time-consuming preprocessing steps, we have unified the input images into npz format with intensity range [0, 255], including five keys
imgs: 3D leison image, shape=(Z,Y,X)gts: 3D lesion mask, shape=(Z,Y,X)spacing: spacing of the raw image, shape=(3,)direction: direction of the raw image, shape=(9,)origin: origin or the raw image, shape=(3,)
Here is the an example script to simulate the RECIST 2D marker from 3D lesion masks.
"""
Simulate RECIST 2D markers from lesion 3D mask
"""
import cv2
import numpy as np
import SimpleITK as sitk
from scipy.spatial.distance import pdist, squareform
input_npz_path = "train_npz/CT_Lesion_MSD_liver_53.npz"
input_npz_data = np.load(input_npz_path, allow_pickle=True)
print(input_npz_data.keys()) # imgs, gts, spacing, direction, origin
gt_array = input_npz_data["gts"]
lesion_ids = np.unique(gt_array)[1:]
RECIST_array = np.zeros_like(gt_array, dtype=np.uint8)
for lesion_id in lesion_ids:
lesion_size = np.sum(gt_array == lesion_id)
if lesion_size <= 1000: # ignore non-measure lesions (rough threshold)
print('Non-measurable lesion with size:', lesion_size)
continue
else:
# get largest 2D slice for the lesion
lesion_array = np.uint8(gt_array == lesion_id)
area_per_slice = np.sum(lesion_array, axis=(1, 2))
key_slice_id = np.argmax(area_per_slice)
largest_2D_slice = lesion_array[key_slice_id, :, :] # key slice to derive the RECIST marker
# get points of the diameter
points = np.column_stack(np.where(largest_2D_slice))
dist_matrix = squareform(pdist(points))
max_diam_idx = np.unravel_index(np.argmax(dist_matrix), dist_matrix.shape)
p1 = points[max_diam_idx[0]]
p2 = points[max_diam_idx[1]]
# generate 2D link marker
cv2.line(img=RECIST_array[key_slice_id, :, :], pt1=np.flip(p1), pt2=np.flip(p2), color=int(lesion_id), thickness=2)
# save the RECIST marker to nifti for visualization
prompt_array_sitk = sitk.GetImageFromArray(RECIST_array)
prompt_array_sitk.SetOrigin(input_npz_data["origin"])
prompt_array_sitk.SetSpacing(input_npz_data["spacing"])
prompt_array_sitk.SetDirection(input_npz_data["direction"])
sitk.WriteImage(prompt_array_sitk, "RECIST_marker_demo.nii.gz")
# save image
img_sitk = sitk.GetImageFromArray(input_npz_data["imgs"])
img_sitk.CopyInformation(prompt_array_sitk)
sitk.WriteImage(img_sitk, "img_demo.nii.gz")
- Downloads last month
- 188